WHO says COVID variants will ‘pick off’ the most vulnerable people. As many different variants of the virus emerge worldwide and threaten global health, detection of the variants becomes particularly important.
Our product DV101: SARS-CoV-2 Nucleic Acid Detection Kit Cy5/ROX (Multiplex Real Time RT-PCR) has been evaluated to detect 15 different SARS-CoV-2 virus variants. An in silico analysis of the primers and probe sets compared with mutations sites of 15 variants, indicates that the mutation sites of the 15 variants are not located in the primer binding regions targeted by the kit. The results ensure the accuracy and sensitivity of the kit remains consistent between the initial SARS-CoV-2 virus strain and the 15 variants, without the risk of drop out or target failure. But it cannot differentiate the variants or identify which variants are present in the sample.
The 15 variants of SARS-CoV-2 are divided into Variants of Concern (VOC), Variants of Interest (VOI) and currently designated Alerts for Further Monitoring. The variants are like below:
Variants of Concern (VOC):
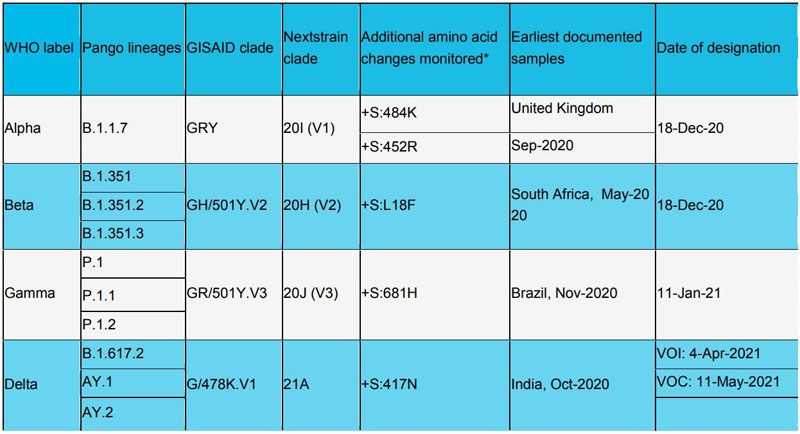
Variants of Interest (VOI):
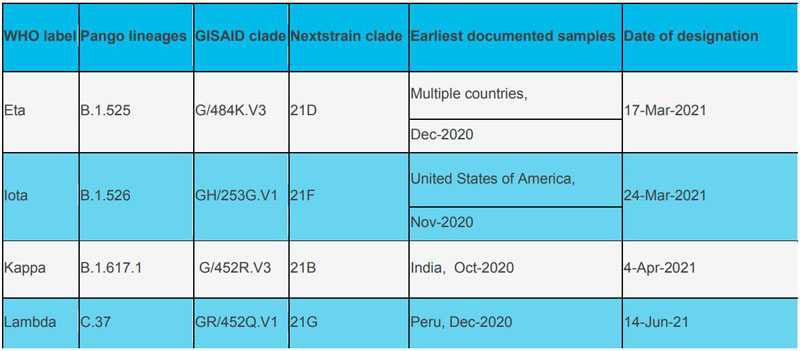
Currently designated Alerts for Further Monitoring:
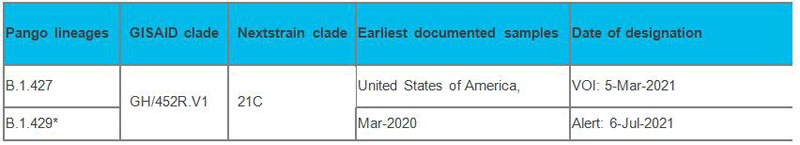
The work methodology we use in the detection of the variants is sequence alignment. The primers and probes of this kit are designed at specific sites of ORF1a, ORF1b and N genes of the SARS-CoV-2 virus. Aligning the sequences of the variants with primer binding regions can determine whether the primers annealing sites are affected. When there is no mismatch between the sequences of the variants and primer binding regions, it proves that the primers can anneal to the specific sites of the variants and amplify the sequences for detection. The sequence alignment tools used are NCBI -BLASTn, Snapgene.exe and USCS In-Silico PCR.
While the in silico assessment of our kit has demonstrated no impact on the accuracy and sensitivity theoretically, all molecular tests may be subject to false negative results due to high sensitivity, and the risk of false negative results may increase when testing genetic variants of SARS-CoV-2 by kits with other primers and probe sets resulting in mismatches with the variants. However, it was shown that the reported negative results were more often due to improper sample collection or collection time, i.e., too soon before symptom onset or too late after symptom release, rather than the reduced specificity of the commercial kits.
Reference:
1. US Food and Drug Administration. Genetic Variants of SARS-CoV-2 May Lead to False Negative Results with Molecular Tests for Detection of SARS-CoV-2 - Letter to Clinical Laboratory Staff and Health Care Providers 2021. Available from: https://www.fda.gov/medical-devices/letters-health-care-providers/genetic-variants-sars-cov-2-may-lead-false-negative-results-molecular-tests-detection-sars-cov-2
2. Mathieu Gand, et al. Deepening of In Silico Evaluation of SARS-CoV-2 Detection RT-qPCR Assays in the Context of New Variants. Genes 2021, 12, 565. https://doi.org/10.3390/genes12040565









