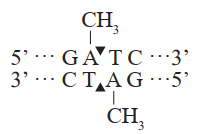This product is an improved version of DpnI restriction enzyme, exhibiting higher activity compared with conventional DpnI enzyme. This enzyme can effectively recognize and cleave the sequence GmATC (A is methylated) and cannot cleave the sequence GATC (A is not methylated). DMT Enzyme is compatible with reaction buffers of multiple PCR enzymes (such as EasyPfu, TransStart, FastPfu, etc.). After PCR, this enzyme can be added directly to the reaction system to start digestion reaction. After digestion reaction, downstream transformation experiments can be performed without heat inactivation.
Restriction Site
Highlights
High efficiency, high specificity, no star activity.
Applications
In vitro site-directed DNA mutagenesis; degradation of methylated plasmid template
Storage
at -20°C for two years
Shipping
Dry ice (-70 ℃)
1.Liu M , Feng Z , Ke H , et al. Tango1 spatially organizes ER exit sites to control ER export[J]. The Journal of Cell Biology, 2017, 216(4):1035-1049.
2.Bingqing L , Yingying Y , Yajie Z , et al. A Novel Enterovirus 71 (EV71) Virulence Determinant: The 69th Residue of 3C Protease Modulates Pathogenicity[J]. Frontiers in Cellular and Infection Microbiology, 2017, 7:26.
3.Wang T N , Zhao M . A simple strategy for extracellular production of CotA laccase in Escherichia coli, and decolorization of simulated textile effluent by recombinant laccase[J]. Applied Microbiology and Biotechnology, 2017, 101(2):685-696.